GraphIBD
Overview
GraphIBD provides the following methods:- Standard Pairwise method
- Pairwise method detects excessive IBD rate in case/case pairs. The standard approach is permutation test (slow).
- Fast-Pairwise method
- Performs pairwise method much faster using a novel importance sampling approach.
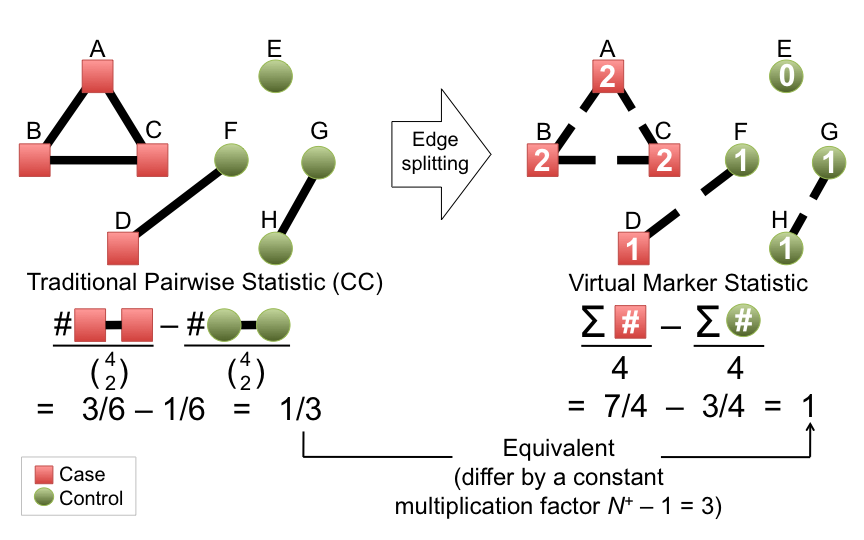
- Virtual markers
- Build virtual marker based on the degree of the vertices. Virtual markers can be directly used for association testing. The virtual marker testing is more flexible than pairwise method in that quantitative traits can be tested and covariates can be included.
Download
GraphIBD version 0.1.0 containing the followings,
GraphIBD.jar (java archive package file)
example.bgl (example input file: Beagle format)
example.bgl.fibd (example input file: Beagle FastIBD output format)
README
LICENSE
Java source code files
Source codes will be available shortly.
Version/bug info
v0.1.0 (2012-12-29) Initial prototype version deployed
User's guide
Option summary:
usage: java -jar GraphIBD.jar [options]
BASIC OPTIONS
-beagle <beagle_file fibd_file> Beagle format file with phenotype row and fastIBD
result file
-single <ibd_file phenotype_file> Single marker IBD information files
-out <file> Output file prefix (default="outfile")
-help Print help
-verbose Print verbosely (default=false)
-seed <INT> Seed for random number generator (default=current
time)
PAIRWISE METHOD OPTIONS
-pairwise Perform pairwise test (default=false)
-pw_method <name> Pairwise test: testing method (CN, CC)
(default=CN)
-pw_sampling_scheme <name> Pairwise test: Sampling scheme (isample, permute)
(default=isample)
-pw_num_sampling <INT> Pairwise test: Number of permutation or isampling
(default=1000)
-pw_adaptive_num_sampling <P1 N1 P2 N2 ...> Pairwise test: Adaptive sampling which, given P
and N pairs, samples N times if p-value is < P
(default=none)
-pw_auto_avg_adjust Pairwise test: Automatically adjust for genomic
average using all SNPs in file (default=false)
-pw_manual_avg_adjust <CASE_ADJ CONT_ADJ> Pairwise test: Adjust for genomic average of cases
and controls with these specified values (default=
0 0)
-pw_no_pvalue Pairwise test: Do not compute p-values, compute
just statistics (default=false)
VIRTUAL MARKER CONSTRUCTION OPTIONS
-vmarker Generate virtual markers (default=false)
-vm_auto_avg_adjust Virtual marker: Automatically adjust for genomic
average of virtual marker (default=false)
-vm_manual_avg_adjust <FILE> Virtual marker: Adjust for genomic average for
each marker with the values in the file
Example Running Command:
java -jar GraphIBD.jar -beagle example.bgl example.bgl.fibd
Output format:
Four columns: RSID, P-value, Case/case IBD rate, Contrasting IBD rate
Publication
Buhm Han, Eun Yong Kang, Soumya Raychaudhuri, Paul I. W. de Bakker and Eleazar Eskin, “Fast Pairwise IBD Association Testing in Genome-wide Association Studies”, Bioinformatics (2013).
Contact
Buhm Han : buhmhan (AT) broadinstitute.org , http:www.broadinstitute.org/~buhmhan
Eun Yong Kang: ekang (AT) cs.ucla.edu
Funding information
B.H., E.Y.K. and E.E. are supported by National Science Foundation grants 0513612, 0731455, 0729049 and 0916676, and NIH grants K25-HL080079 and U01-DA024417. B.H. is supported by the Samsung Scholarship. This research was supported in part by the University of California, Los Angeles subcontract of contract N01-ES-45530 from the National Toxicology Program and National Institute of Environmental Health Sciences to Perlegen Sciences.