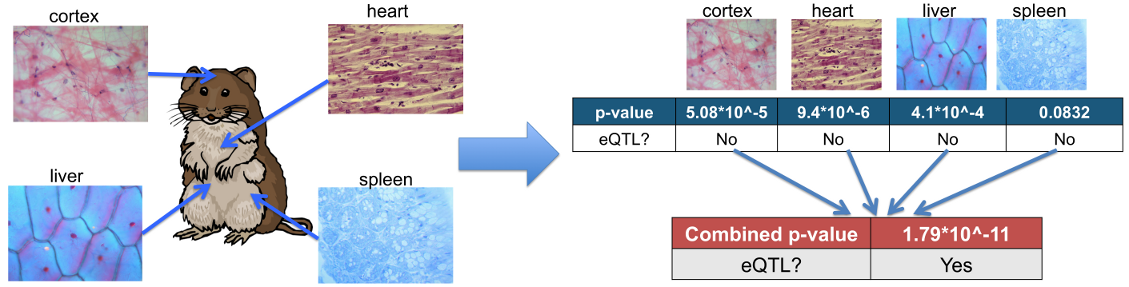
Instruction step 2: Convert Multiple Tissue Dataset to Meta-Tissue format
Once users prepared multiple tissue dataset discussed in the previous step (Step 1), they need to convert the dataset into the format for Meta-Tissue. For this, we provide a Java program called "MetaTissueInputGenerator" and here is its usage. #1 means the first item in Step 1.
|
Here is a sample command using the input files in "example/1_orig_input/" folder and using "example/2_MetaTissue_input/" as output folder in the software package.
|
IMPORTANT!!! Please use the full or absolute path (e.g. /usr/home/[user_id]) rather than using relative path (e.g. "../") or tilde ("~").
IMPORTANT 2!!! Please do not edit or change the output files (output_gene.txt, output_snp.txt, and matrix.txt files above). Manually creating the output files or changing them will cause errors and unexpected behaviors.
Since v0.4, Meta-Tissue supports covariate file. Please use -d and -s options to create covariate file needed for the next step.
Since v0.4, Meta-Tissue supports dosage in genotype data (such as dosage from imputation). To use dosage data, one needs to replace genotype file specified by -a option in MetaTissueInputGenerator with a genotype file that contains dosage information and use -x option. The file format of the genotype file with dosage information is that each line is each SNP, and each column is dosage of each individual (one value). If there are 3 probabilities available for each imputed genotype such as P(A1/A1), P(A1/A2), and P(A2/A2), one can compute
P(A1) = 2*P(A1/A1) + P(A1/A2)
and use this dosage for Meta-Tissue. IMPORTANT!!! The order of individuals in this genotype file must match the order of individuals specified in the individual file (-b option). Below is an example of dosage genotype file that includes 10 SNPs and 5 individuals.
0.769 0.799 1.782 0.252 0.216
1.213 1.672 0.139 1.198 0.81
0.707 1.51 1.002 0.031 1.661
0.557 0.881 0.609 0.361 0.277
0.153 0.639 1.768 0.788 0.159
1.01 1.529 1.162 0.903 0.526
1.459 0.414 1.575 0.696 1.669
1.509 1.377 1.75 0.049 0.755
0.931 0.112 0.608 1.164 1.463
0.079 0.849 0.785 0.981 0.164
P(A1) = 2*P(A1/A1) + P(A1/A2)
and use this dosage for Meta-Tissue. IMPORTANT!!! The order of individuals in this genotype file must match the order of individuals specified in the individual file (-b option). Below is an example of dosage genotype file that includes 10 SNPs and 5 individuals.
0.769 0.799 1.782 0.252 0.216 1.213 1.672 0.139 1.198 0.81 0.707 1.51 1.002 0.031 1.661 0.557 0.881 0.609 0.361 0.277 0.153 0.639 1.768 0.788 0.159 1.01 1.529 1.162 0.903 0.526 1.459 0.414 1.575 0.696 1.669 1.509 1.377 1.75 0.049 0.755 0.931 0.112 0.608 1.164 1.463 0.079 0.849 0.785 0.981 0.164