Instruction step 4: Run Metasoft (Meta-analysis software)
The last step in Meta-Tissue is to run software called Metasoft that performs meta-analysis. We already provided the script that runs Metasoft in Step 3. Hence, all users need to do is to execute the script.
[output prefix].SNP.[start_SNP_index].metasoft.sh
1. If users parallelized MetaTissueMM in Step 3, then users also need to parallelize this step as well; run this shell script for each [start_SNP_index] file.
2. Running this script will generate a following output file:
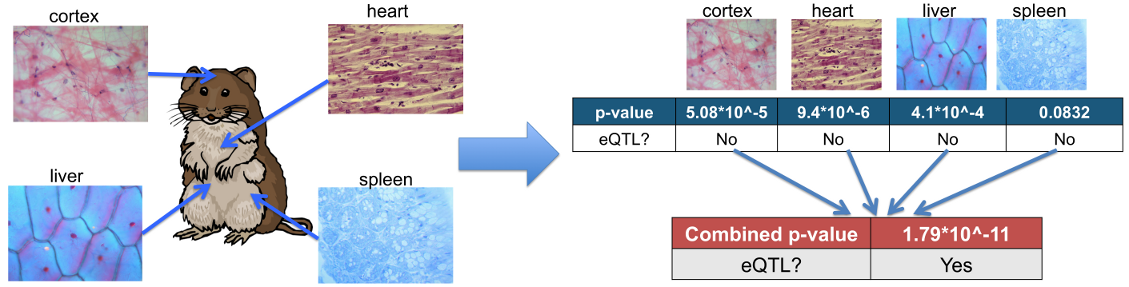
Each line in output file contains p-value for each pair of SNP and gene expression from meta-analysis that combines information from multiple tissues; hence one p-value for each pair of SNP and gene expression pair. The first column indicates ID for a pair of SNP and gene expression, and ID is "[SNP ID]:[Probe ID]"
|